what does dna polymerase 3 need in order to attach to the dna template
PCR stands for Polymerase Concatenation Reaction which is ane of the fundamental methods of molecular biology.
Substantially, the primary purpose of polymerase concatenation reaction is to rapidly increment the number of copies of specific Deoxyribonucleic acid regions.
Information technology consists of 3 basic PCR steps and a relatively complex reaction mixture.
The product of the polymerase chain reaction acts equally the means of further analysis. For instance, PCR is used along with gel electrophoresis to detect different Dna sequences.
Today, different types of PCR technique, combined with other technologies, find numerous applications in such fields equally research, forensic science, agricultural sciences, medicine, etc. Information technology is used to diagnose diseases, clone and sequence genes.
Polymerase concatenation reaction can be performed using DNA from a variety of sources. It is very sensitive and needs but trace amounts of nucleic acids to produce enough copies for conventional laboratory assay.
The polymerase chain reaction procedure serves to raise the number of Deoxyribonucleic acid fragments

three basic steps of PCR process
The polymerase concatenation reaction is a three step cycling process consisting of defined sets of times and temperatures.
3 basic PCR steps include:
- denaturation step;
- annealing stride;
- extension (elongation) step.
Each of these polymerase concatenation reaction steps is repeated 30–40 times (cycles).
In the grade of each wheel, the PCR reaction mixture is transferred between three temperatures.
Profile of iii bones PCR steps
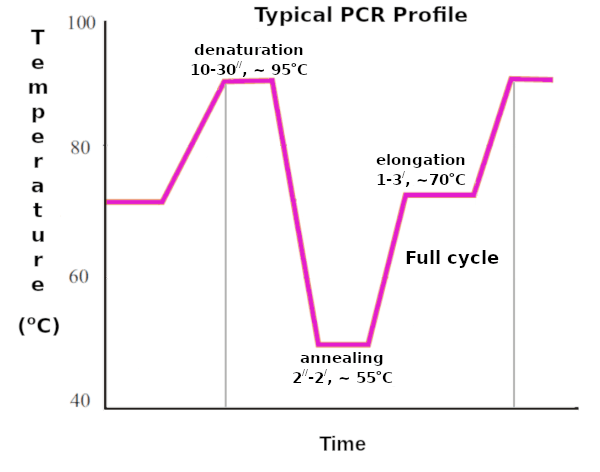
During successive cycles of basic PCR steps (denaturation, annealing, and extension) all the new strands will act every bit DNA templates causing an exponential increase in the amount of Deoxyribonucleic acid produced.
Each bicycle doubles the number of Deoxyribonucleic acid molecules (amplicons) amplified from the Dna template.
Starting time polymerase chain reaction step – DNA denaturation
The outset of iii PCR steps is a denaturation pace.
During the denaturation step, the hydrogen bonds that hold together the two strands of the double-stranded nucleic acids are broken and the strands unwind from each other.
This process releases single-stranded DNA to act as templates in the last PCR extension footstep.
The denaturation temperature is in a higher place 90°C (usually 94°C) and the time is up to ane infinitesimal (usually xxx seconds).
Polymerase chain reaction steps

Second polymerase concatenation reaction step – DNA Primer annealing
At the annealing step, DNA primers line up on exposed nucleotide sequences at the Deoxyribonucleic acid target according to base-pairing rules. This is a typical temperature-dependent DNA : DNA hybridization reaction and has to be optimized.
This is the just temperature in a PCR cycle steps that can be widely varied.
The temperature depends on the verbal sequence and length of the primers.
Commonly, the PCR reaction mixture is cooled down to 40–60°C. Notwithstanding, annealing temperatures for DNA templates with a high GC content tin can be equally high as 72°C (the normal temperature of the extension step).
The temperature for this PCR stride is called for the optimum binding of the Deoxyribonucleic acid primers to the correct DNA template and depends on primer'south melting temperature. The wrong annealing temperature can result in simulated products, or in no detectable products at all.
Since the primers are relatively curt, and at high molar concentrations, duration of the annealing footstep is around xxx seconds.
At this stride, the annealed oligonucleotides provide a gratis three' hydroxyl grouping for Taq polymerase and act as primers for synthesis of nucleic acids.
Final polymerase concatenation reaction step – DNA synthesis
The concluding of 3 basic PCR steps is called extension or elongation step.
Information technology is the Deoxyribonucleic acid synthesis step and carried out by a thermostable Deoxyribonucleic acid polymerase (normally Taq polymerase).
The temperature of the elongation step is unremarkably set at 72°C. Information technology is slightly below the optimum for Taq polymerase.
The Taq polymerase produces complementary DNA strands starting from the primers.
The synthesis gain at approximately grand bases per minute.
Therefore, to amplify a Deoxyribonucleic acid template that is 500 bases in length, under normal weather a time of the PCR extension stride should be at to the lowest degree 30 seconds.
Commonly, PCR extension time is xxx seconds for every 500 bp (base of operations pair) of product.
DNA amplification process during PCR cycles
During the very outset PCR wheel the only templates available for primer annealing are the target nucleic acids.
Because the initial template is many times larger than the length of the desired amplicon, the polymerization of the first wheel will proceed until it is interrupted at the denaturation step of the second wheel.
At the finish of the first PCR bike, in that location are ii double-stranded nucleic acid molecules for each 1 that the reaction started with.
Each nucleic acid molecule contains one strand of the original template, and one novel strand, which is bounded at one cease by the oligonucleotide primer and at the other end by how far polymerization was able to keep during the extension step.
These PCR products grade Dna templates that are bounded on only one finish (semi-bounded DNAs).
In the second bicycle, both the original nucleic acrid targets and the semi-bounded DNAs will serve as templates.
Original DNA templates will proceed to make semi-bounded products in every cycle of the polymerase chain reaction.
Starting with the second cycle of PCR distension, semi-divisional DNAs will class the PCR amplicons.
In every subsequent cycle, the DNA templates, the semi-bounded DNAs, and the amplicons volition serve as templates for the PCR primers.
In a PCR reaction mixture:
- the corporeality of template Deoxyribonucleic acid does not change;
- the number of semi-bounded Dna templates increases arithmetically every cycle;
- every cycle starting with cycle 2, the number of amplicons increases geometrically.
At the end of 35 PCR cycles at that place are more than 34 billion copies of the DNA amplicons for every copy of the original template DNA sequence.
The development of the programmable thermocycler helped spread the new PCR engineering science.
The programmable thermocycler is based on metal heating blocks with holes for the PCR tubes and designed to switch between the programmed series of temperatures of polymerase chain reaction steps.
Related manufactures
References & further readings:
- Variants of PCR – Wikipedia
- PCR Protocol for Taq DNA Polymerase with Standard Taq Buffer – New England Biolabs
- PCR Technique with its Application – Enquiry & REVIEWS - INTERNATIONAL JOURNALS
- PCR Protocols & Applications – QIAGEN
- PCR Cycling Parameters—Six Primal Considerations for Success – Thermo Fisher Scientific
Source: https://biology.reachingfordreams.com/biology/molecular-genetics/methods-in-molecular-genetics/20-3-pcr-steps-of-dna-amplification
0 Response to "what does dna polymerase 3 need in order to attach to the dna template"
Post a Comment